Do you want to keep up to date? Subscribe to our newsletter. 1mail every 2months! |
 |
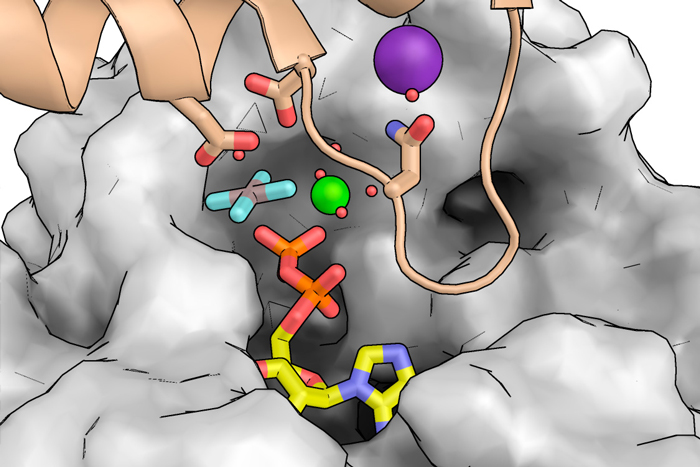
1st April 2022. Antibiotic resistance causes more than one million deaths a year worldwide and it is estimated that this figure could increase tenfold in the coming decades. About one-tenth of current deaths are due to infections caused by methicillin-resistant S. aureus infections. The FtsZ protein of this pathogen, the subject of the study, plays a central role in cell division, which is necessary for the spread of these infections.
"The activity of FtsZ in the bacterial division makes it a target for the discovery of new antibiotics," says Federico M. Ruiz, lead author of the study. "FtsZ forms filaments that, in the presence of cations, hydrolyze the nucleotide GTP. These filaments grow and decay simultaneously at opposite ends, causing a movement similar to that of a treadmill. Thus, in association with other proteins, FtsZ forms a ring that allows the remodeling of the bacterial cell wall and leads to cell division".
This research, co-directed by Carlos Fernández-Tornero and José Manuel Andreu from the Margarita Salas Center for Biological Research (CSIC), has been recently published in the journal PLoS Biology.
Using X-ray crystallography at the ALBA Synchrotron and the European Synchrotron Radiation Facility (ESRF) in France, researchers have obtained high-resolution structures of FtsZ filaments from S. aureus in complex with different cations and nucleotide analogs, including those that mimic the ground and transition states of catalysis. The structures provide atomic details of the process by which FtsZ coordinates GTP hydrolysis with conformational changes essential for the dynamics of the filaments formed by this enzyme. Combined with mutational and biochemical analysis, the structures obtained at XALOC beamline and ID23-1 allowed the identification of FtsZ amino acids involved in this process.
"All together —concludes Sonia Huecas, a member of the research team— these findings provide insight into the intimate mechanism of bacterial cell division and open up a pathway for biomedical applications".




