ALBA Synchrotron
Using synchrotron light, researchers from CIC bioGUNE have solved the structure of RavN, a protein that Legionella pneumophila uses for stealing functions and resources of the host cell.
Cerdanyola del Vallés, 6th July, 2018.
Mimicry is the ability of some animals to resemble others in their environment to ensure their survival. A classic example is the stick bug whose shape and colour make him unnoticed to possible predators. Many intracellular pathogens also use molecular mimicry to ensure their survival. A part of a protein of the pathogen resembles another protein totally different from the host and many intracellular microorganisms use this capability to interfere in cellular processes that enable their survival and replication.
The Membrane Trafficking laboratory of the CIC bioGUNE in the Basque Country, led by Aitor Hierro, in collaboration with other groups from the National Institutes of Health in the United States, have been working for several years in understanding how the infectious bacterium Legionella pneumhopila interacts with human cells. During this research, experiments have been carried out at the and I04 beamline of Diamond Light Source (UK). The results enabled scientists to solve the structure of RavN, a protein of L. pneumophila that uses this molecular mimicry to trick the infected cell.
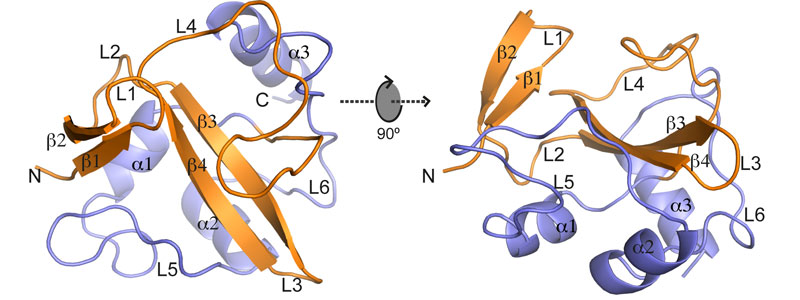
During this study, researchers have proved how Legionella uses molecular mimicry to exploit ubiquitination, a process that modifies proteins containing ubiquitin for their participation in different cellular processes. Three types of enzymes are taking part in ubiquitination: activation (E1), conjugation (E2) and finally ubiquitin ligases to the target protein (E3). Using X-ray crystallography, the structure of the RavN protein of L. pneumophila has been determined, showing that it has a certain structural similarity with E3 eukaryotes despite its different composition. Based on this result, a secondary homology search was carried out with other Legionella proteins and they discovered new E3 ligase candidates that hadn’t been identified up to date. The work shows that L. pneumophila has a group of E3 ligase enzymes that are vital for the infection process and despite the evolutionary divergence, the regions of molecular mimics are highly conserved to facilitate take possession of the host cell ubiquination process.
What is Legionellosis?
The causal agent of Legionellosis is a Gram-negative bacterium of the Legionella genus whose natural habitat is the stagnant water areas where it lives. But also the artificial systems of water storage such as outdoor fountains, swimming pools, hot water tanks and air conditioning cooling towers without proper maintenance are also favourable environments for their growth and become the main sources of infection. The transmission of Legionella is commonly by inhalation of water droplets or polluted water sprays.
Legionella was discovered in 1977 following an outbreak of pneumonia at the 1976 American Legion Convention in Philadelphia, USA, and hence the name Legionnaires' Disease or Legionellosis. Legionella infection ranges from mild flu-like conditions (Pontiac fever) to life-threatening forms of pneumonia, especially in the elderly, and people with previous medical problems. According to a study developed by the European Centre for Disease Control (ECDC), between 2011 and 2015, 30.532 cases of Legionellosis were reported, 70% taking place in Germany, Spain, France or Italy.

Figure: Schematic representation of the structure of RavN1-123 as ribbon diagram displayed in two orientations (rotated by 90° along the x axis). Secondary elements are indicated as spirals (helices) or arrows (beta strands), with the RING/U-box motif colored in orange and the C-terminal structure colored in slate.