ALBA Synchrotron
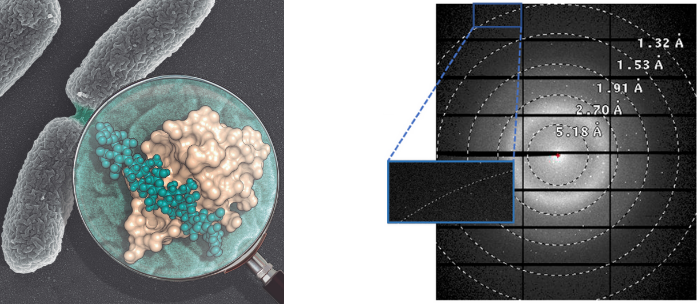
Their high division rate is one of their main weapons for antibiotic resistance. This work opens the door to the development of drugs that can block this precise mechanism. Some of the experiments have been made in the XALOC beamline at ALBA.
The high division rate of bacteria, that is, the successful way in which they reproduce, is one of their best weapons against antibiotics. A team from the Rocasolano Institute of Physical Chemistry (IQFR-CSIC) in Madrid, Spain, the University of Notre Dame in the United States and the National Centre for Biotechnology (CNB-CSIC) in Madrid, Spain, has revealed the structure of a key machinery in the process of bacterial division. The conclusions, published in the latest issue of the journal Nature Communications, open the door to the design of a future drug capable of blocking this precise machinery, without which the bacteria become sensitive to the antibiotic effect.
A wide group of various proteins that assemble in an ordered and dynamic way orchestrates the bacterial cell division. Thus forming a precise machinery that guarantees the reproductions proper development. Virtually all bacterial species have specialized domains that recognize the bacterial wall (composed of peptidoglycan) at the time of division and allow the correct location in space and time of these proteins during the generation of the two daughter cells from the mother cell.
The scientists have used the X-ray crystallography technique, in the XALOC beamline at ALBA, to obtain the structure of these specialized domains. More specifically, they have studied the SPOR (Sporulation-related repeat) domain of the RlpA protein of Pseudomonas aeruginosa, a multiresistant bacteria. There are few antibiotics available against it, considered by the World Health Organization as a "critical priority pathogen".
"The SPOR domains are present in almost all bacteria during the division process. However, despite their relevance, until now nobody had been able to explain how they work during cell division. All of these proteins recognize the same type of cell wall produced during bacterial division. Our work unveils, for the first time, why all SPOR domains recognize the same type of wall to facilitate the division", explains Juan Antonio Hermoso, a researcher at the Rocasolano Institute of Physical Chemistry.
The research suggests a model, comparably to all kinds of bacteria. "Our results provide information at the atomic level on how the SPOR domains get connected to the bacterial cell wall and, thus, open the door to the development of molecules that can specifically block these domains, which would make bacteria sensitive to antibiotics", highlights Hermoso.
Link to CSIC’s news: https://www.csic.es/es/actualidad-del-csic/investigadores-del-csic-revelan-la-estructura-de-una-maquinaria-clave-en-la
With the collaboration of Fundación Española para la Ciencia y la Tecnología. The ALBA Synchrotron is part of the of the of UCC network from the FECYT and has received support through the FCT-20-15798 project.